CRISPR : Clustered Regularly Interspaced Short Palindromic Repeats , is a bacterial immune system that has been modified for genome engineering.
The name, while long tells a lot about the CRISPR system. “Repeats”, like the name implies are “short” repeats of DNA segments typically about 20 - 40 letters in length. There also “Palindromic”, meaning that the sequence of letters can be read the same both left to right or right to left.

Now these “repeats” are “interspaced”. In between these repeats we have what’s called spacer DNA. Unlike the repeats, spacer DNA is not identical, each spacer is unique. These spacer segments are actually viral \ bacteriophage DNA.
There are also CRISPR Associated genes called CAS genes. These genes code for CAS proteins mainly helicases (used to unzip or unwind DNA) and nucleases (which cut DNA).

The theory is that bacterium like E.Coli use the CRISPR system as an immune system response against foreign threats like viruses. Essentially, what happens is that CAS complex will find a matching segment in the DNA and use it to identify a foreign virus RNA segment, this is called the CRISPR RNA (or crRNA). When the complex finds the matching RNA, it simply just destroy’s it and the threat is removed.
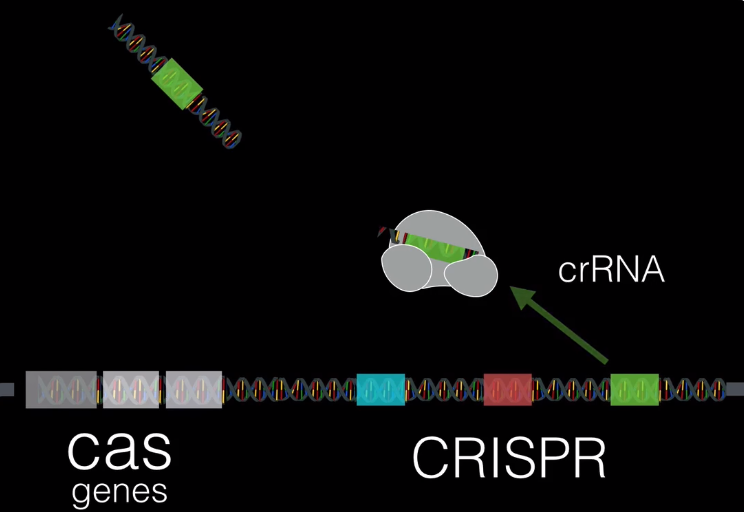
If, on the other hand the viral RNA does not match with anything in the bacteria’s genome, the CRISPR system will create a different protein, known as a class I CAS protein. This system, very simply, takes the RNA from the virus, breaks it apart and and copies it into the CRISPR system, creating a new spacer. Essentially, these spacers in bacterial DNA act as a historical repository for old viral infections.
This system was researched by two scientists, Jennifier Doudna and Emmanuel Charpenter. Through the research of their team they were able to show that the CAS9 protein complex (from Streptococcus Pyogenes) could be used to essentially just cut DNA segments. In the bateria the researchers were working on Streptococcus Pyogenes used a complex called CAS9. The cas9 complex uses both tracrRNA and crRNA as well as having two nucleotides used to cut DNA.
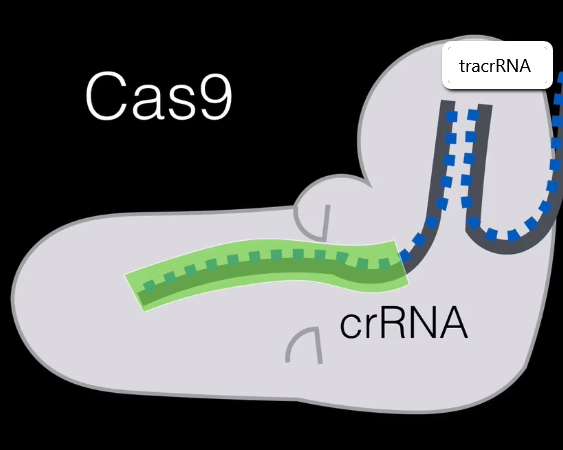
Looking at the cas9 we can see that we still have the spacer RNA (which is the part that will correspond to the matching viral RNA) and to the right we have the tracrRNA, holding the crRNA in place. This whole complex is used to cut DNA.
The team of researchers decided to use the one cas9 protein but put a custom sequence of RNA where the crRNA is. Then they connected the tracrRNA together with the crRNA, called the guide RNA (gRNA). This system would allow the complex to activate and using the gRNA find its target in any DNA strand.
Once cut (called a double strand cut) the body tries to do a couple of things. Using a pathway called non-homologous end joining (NHEJ), the cell can repair the DNA by knowing where the issue is. The problem however, is that the cell doesn’t know if any DNA was actually removed or added or modified just that something is wrong in that area and that it needs to be repaired.
so either it will add mutations into the the cut space and try and regenerate the cut out segemnt (performs insertions/ deletions). These insertions or deletions typically cause the gene to reach the STOP codon early making the gene useless or end later producing an improper protein. In either of these cases the resulting gene would simply not work and this is what’s known as a knockout mutation.
Another result would be that the seperated DNA repair process combines the seperate ends together.
Finally, and most interestingly, the repair process may try and look for the missing base pairs by looking at copies that were made in the past (usually the cell duplication process) by finding a copy of the genome and using that to repair the original. This means however that is possible to add a host RNA (provided by us) that the DNA can use to repair the missing base pairs.
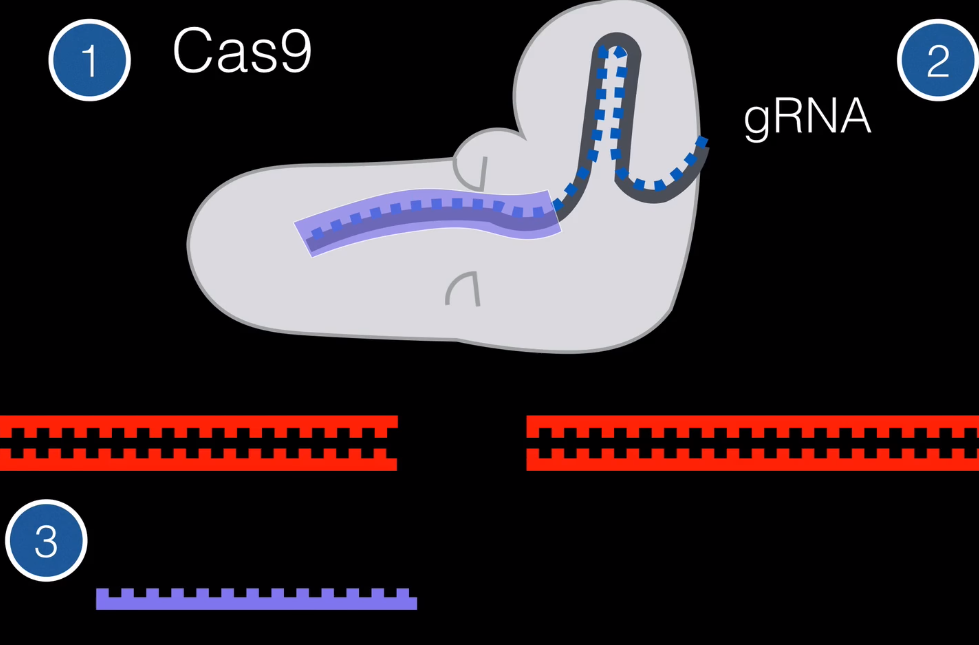
So simply put, we have three parts. The Cas9 system to do the cut. The gRNA to locate and match an area in DNA. And the host RNA that is used to repair the cut DNA.
gRNA : Short synthetic RNA composed of a “scaffold” sequence necessary for Cas9-binding and a user-defined ∼20 nucleotide “spacer” or “targeting” sequence which defines the genomic target to be modified.